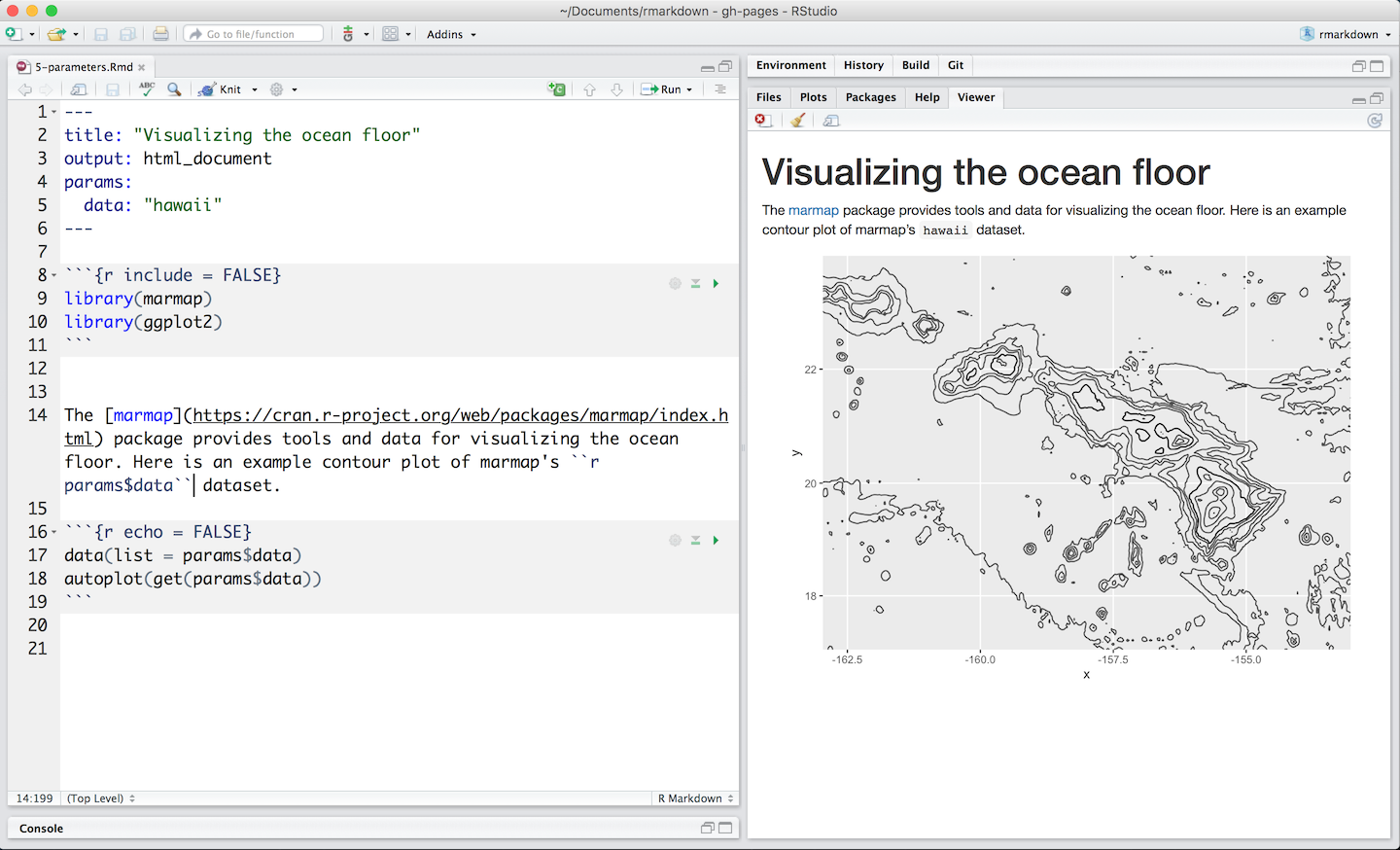
Reproducible Reporting
with Quarto
![]()
Community Engaged Data Science
College of the Atlantic
Acknowledgements


R/Medicine Data Cleaning 2023 Workshop taught by Crystal Lewis, Shannon Pileggi, and Peter Higgins
ASA Traveling Courses on Quarto taught by Mine Çetinkaya-Rundel and Andrew Bray
Disclaimer and license
Opinions expressed are solely my own and do not express the views of my employer or any organizations I am associated with.
This work is licensed under Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International (CC BY-NC-SA).
Jadey Ryan
Data scientist at WA Dept of Agriculture
The Coding Cats: cat & code themed merch


Participation
Log in to Posit Cloud work space:
https://posit.cloud/spaces/504211/content/8093007
If Posit Cloud doesn’t work, download materials locally:
Workshop structure: presentation, 💃🏻 demos, 💪🏻 exercises
💪🏼 Exercise 0
Student introductions:
Your name
Briefly describe your community project & what you’re hoping to learn from this workshop
Community partner // Maine UseR introductions:
Your name, position title, and affiliation
Briefly describe how you use R Markdown or Quarto
15:00
Learning objectives
Learn what Quarto is and what you can use it for.
Learn how to weave code and text together to create a fully reproducible report.
Learn how to use parameters to create variations of a report.
Syntax and RStudio aside
Pipes
2014+ magrittr pipe
%>%2021+ (R \(\geq\) 4.1.0) native R pipe
|>
Isabella Velásquez’s blog post Understanding the native R pipe |> (2022)
To change shortcut to the native pipe:
Tools → Global Options → Code → Editing → Use Native Pipe Operator
Windows: Ctrl + Shift + M
Mac: Cmd + Shift + M
Slide adapted from R/Medicine Data Cleaning 2023 Workshop
Namespacing
package::function()
dplyr::select()
tells R explicitly to use the function
selectfrom the packagedplyrhelps avoid name conflicts (e.g.,
MASS::select())does not require
library(dplyr)
Slide adapted from R/Medicine Data Cleaning 2023 Workshop
RStudio options
Tools → Global Options →
Fussy YAML indentation:
Code→Display→Indentation guides:→Rainbow lines
Match parentheses:
Code→Display→Indentation guides:→ CheckUse rainbow parentheses
Matching divs:
R Markdown→Advanced→ CheckUse rainbow fenced divs
R Markdown → Quarto
R Markdown is an authoring framework supported by many R packages
Figure from “Hello, Quarto” keynote by Julia Lowndes and Mine Çetinkaya-Rundel, presented at RStudio::Conf(2022).
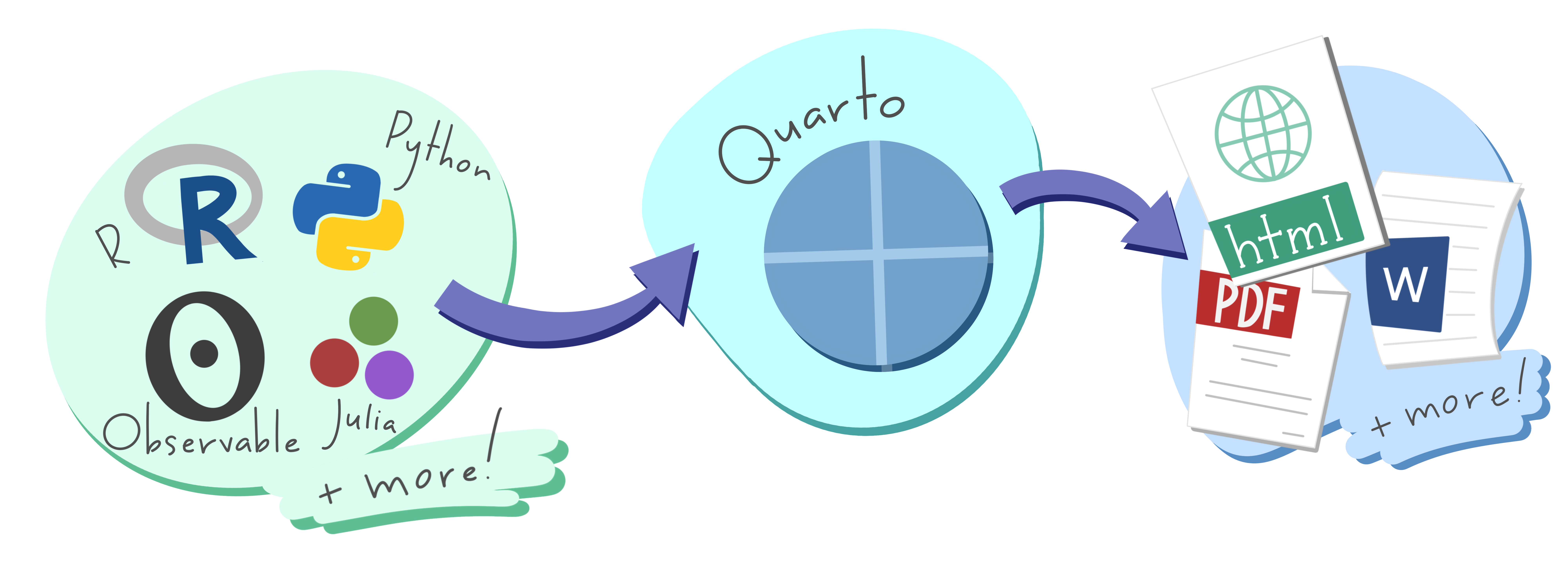
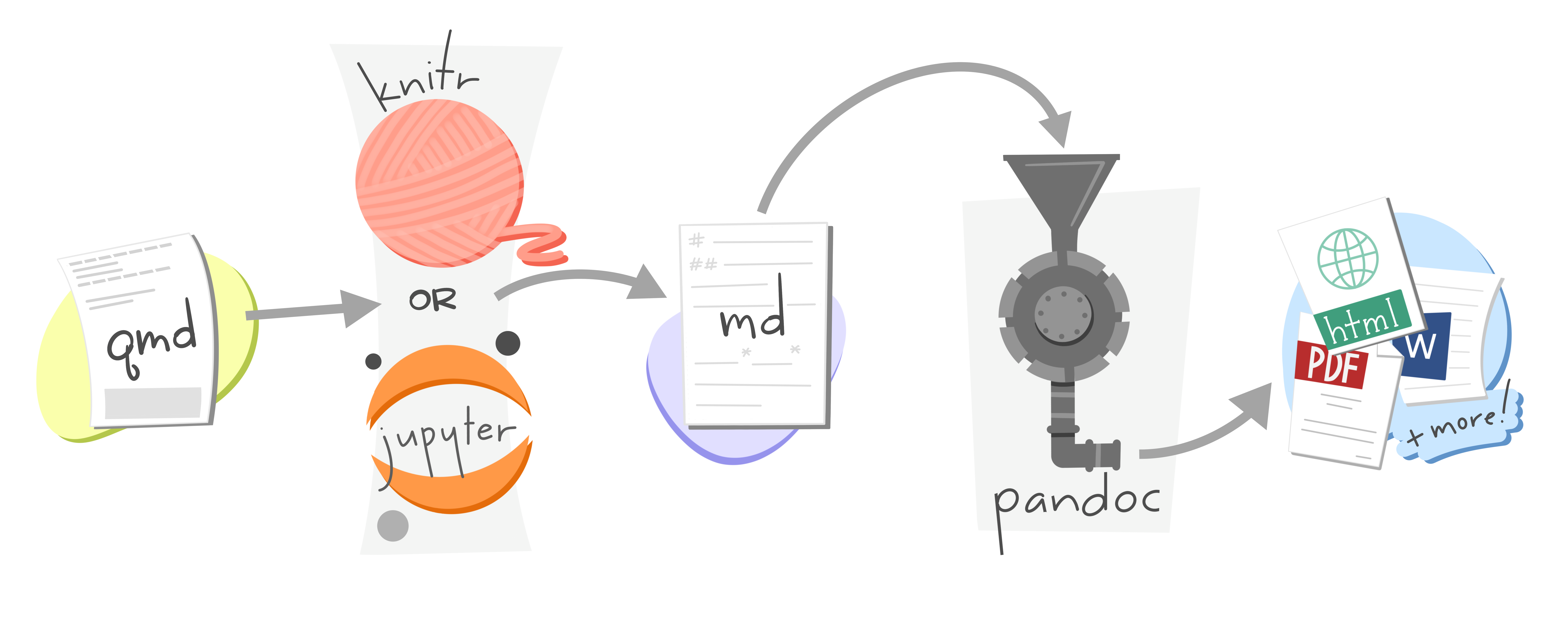
Quarto is a publishing system that supports many multiple & outputs
R Markdown vs Quarto

R Markdown
Vast R Markdown ecosystem
Dependent on R

Quarto
Command line interface (CLI)
Expands R Markdown ecosystem
“Batteries included”
Multi-language and multi-engine
If you’re happy with R Markdown and it’s not broken, no need to switch!
R Markdown will still be maintained but likely no new features (Xie 2022).
How I use Quarto




More real-world applications of Quarto
Convert .Rmd → .qmd
- Change file extension from
.Rmd→.qmd - Change YAML header (
output:→format:) - Convert chunk headers with
knitr::convert_chunk_header()
🆕 Shiny app to convert R Markdown to Quarto ✨
Resources for R Markdown users
From R Markdown to Quarto workshop taught by Dr. Mine Çetinkaya-Rundel and Dr. Andrew Bray.
Ted Laderas’ talk Quarto / R Markdown - What’s Different?
Get started with Quarto
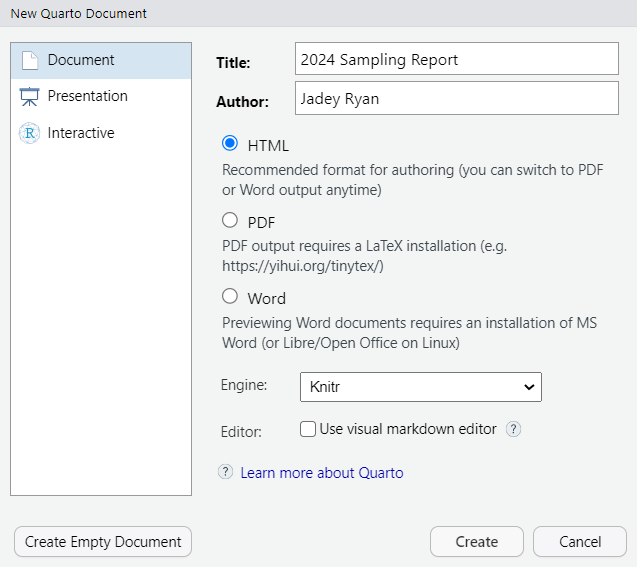
Create a single Quarto document
One .qmd file for a report, presentation, or dashboard
File > New File > Quarto Document…
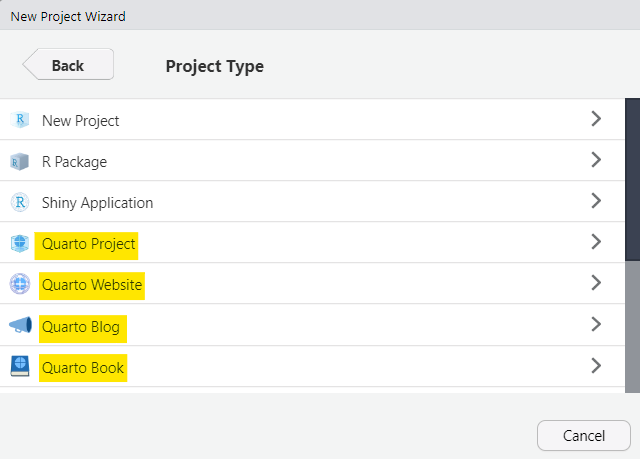
Create a Quarto project
Multiple .qmd files for a website, blog, or book
File > New Project > New Directory > Quarto Project/Website/Blog/Book
Authoring
Toggle between Visual and Source modes with Cmd/Ctrl + Shift+ F4.
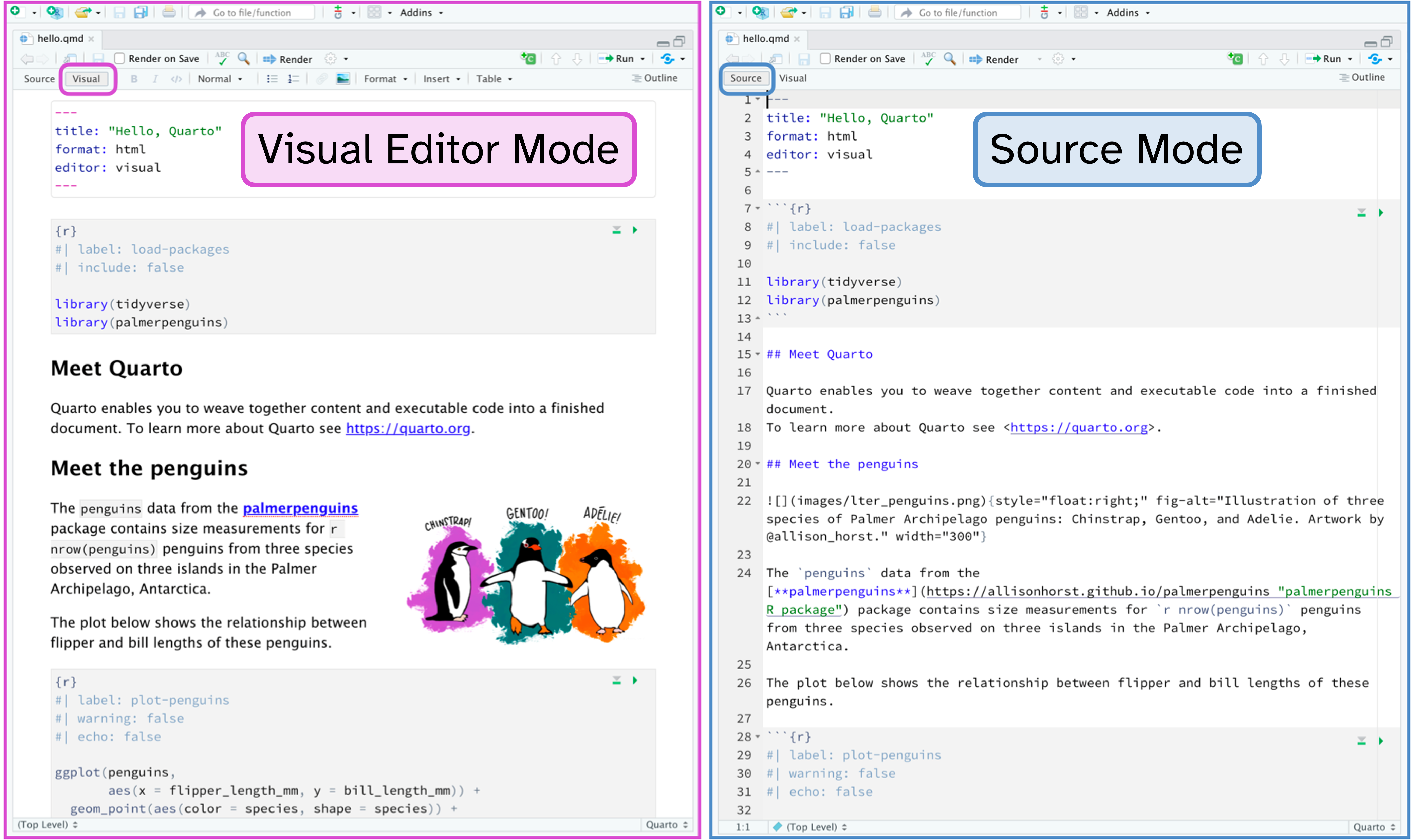
Image adapted from Tutorial: Hello, Quarto.
💪🏼 Exercise 1-1
Create a Quarto document that will generate an HTML output format. Name it
1-hello-quarto.Toggle between Visual and Source modes. What differences in the RStudio IDE do you notice?
In Visual mode, try a traditional keyboard shortcut to format text in Google Docs or Microsoft Word (
Cmd/Ctrl+Bto bold text).Try out other formatting shortcuts you normally use.
Try inserting an image using the Insert Anything shortcut (
Cmd/Ctrl+/).
05:00
Three ways to render
- Use the RStudio/Quarto integration
![]() Render button to render the file and preview the output.
Render button to render the file and preview the output.
Keyboard shortcut: Cmd/Ctrl + Shift + K
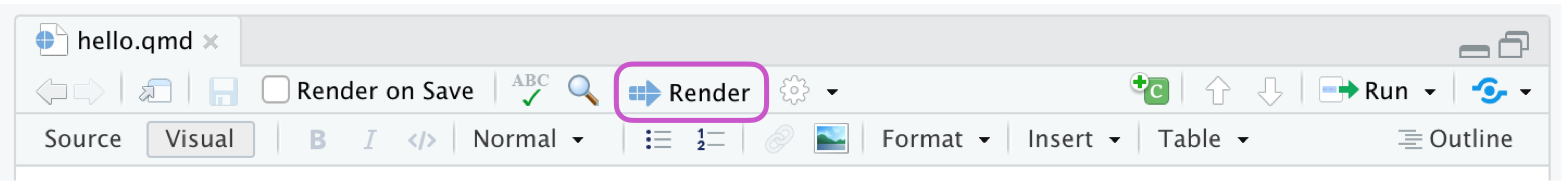
Check the Render on Save option to automatically render and update the preview after saving.
Images from Tutorial: Hello, Quarto.
Three ways to render
- Use the
quartopackage in the Console or an R script
💪🏼 Exercise 1-2
Render the document using the
![Quarto render button in RStudio]() Render button.
Render button.Modify the text and code, render using the
quartopackage, and review the output.Check the Render on Save option, make more changes, save, and watch the preview update.
03:00
Anatomy of a .qmd file
YAML header (metadata and document options)
Narrative (text)
Code chunks (import, wrangle, visualize data)
1. YAML header
“Yet Another Markup Language” or “YAML Ain’t Markup Language”
1---
2title: 2024 Sampling Report
author: Jadey Ryan
date: 2024-04-25
format:
3 html:
theme: flatly
toc: true
---- 1
-
Demarcated by three dashes (
---) on the top and bottom - 2
-
Document level metadata and options using key-value pairs in the format
key: value - 3
- Use 2 spaces for each indentation level – YAML is very fussy!
YAML intelligence: tab-completion
Start a word and then Tab to complete.
Cmd/Ctrl+Spaceto see all available options.
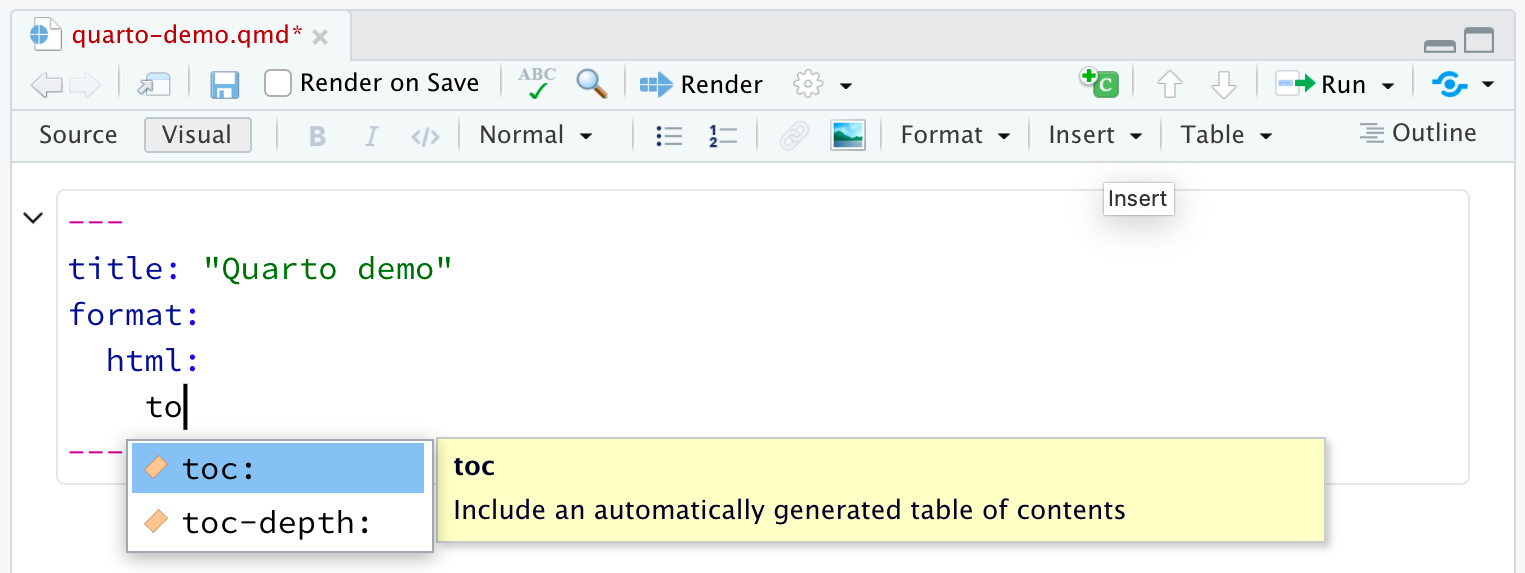
Content and image adapted from YAML Intelligence on quarto.org.
YAML intelligence: diagnostics
Incorrect YAML is highlighted when documents are saved:
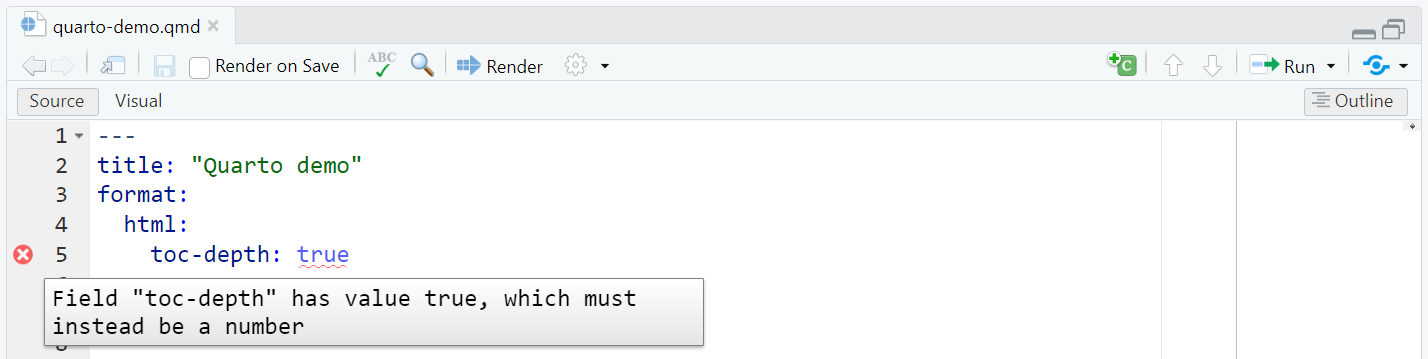
Content and image adapted from YAML Intelligence on quarto.org.
💪🏼 Exercise 1-3
Explore the Quarto documentation on HTML theming and choose a Bootswatch theme.
Add your chosen theme to the YAML.
Cmd/Ctrl+Spaceto see all available YAML options for the HTML format.Try any that sound interesting.
💬 Discuss: Share your two favorites.
Use the HTML reference as needed.
08:00
2. Narrative
Markdown syntax for:
Text with formatting:
**bold**→ boldSection headers:
# Header 1,# Header 2Hyperlinks:
[google.com](https://google.com)→ google.comImages:
Inline code:
2024-04-25→2024-04-25Inline math:
`$E = mc^{2}$`→ \(E = mc^{2}\)
💪🏼 Exercise 1-4
Enable the Visual mode.
Explore the Format, Insert, and Table drop downs.
Pick one or two options from these drop downs and try them out.
Switch to the Source mode and see what that formatting or feature looks like in markdown syntax.
💬 Discuss: Share your two favorites.
05:00
3. Code chunks (or cells or blocks)
Three ways to insert code chunks:
Keyboard shortcut
Cmd/Ctrl+Option/Alt+I.![Insert Chunk button in RStudio]() Insert Chunk button in the editor toolbar.
Insert Chunk button in the editor toolbar.Manually type the chunk delimiters
```{r}and```.
Two ways to run code chunks:
Use the Run Current Chunk or Run All Chunks Above buttons.
![Code chunk in RStudio with the Run All Chunks Above and Run Current Chunk buttons highlighted and labelled.]()
Run the current code chunk with
Cmd/Ctrl+Shift+Enter.
💪🏼 Exercise 1-5
Insert a code chunk at the beginning of the document that includes
library(ggplot2).
Do not run this chunk yet.Insert a chunk at the end that includes
ggplot2code to generate a plot of your choice.
Browse the various geoms on the ggplot2 documentation website for examples.Try running just this plot code chunk. It should error with
could not find function "ggplot". 💬 Discuss: We attached theggplot2package, why can’t R find theggplotfunction?Click the Run All Chunks Above button.
💬 Discuss: Why do you think it worked this time?
08:00
Optional code chunk labels
Use a hashpipe (#|) to specify labels.
Improve documentation and navigation.
Optional code chunk options
Use a hashpipe (#|) to specify options.
Control code execution, text or plot output, layout, captions, etc.
eval: falseprevents code from being evaluated and does not generate results. Use this to display example code or disable a code block.echo: falseprevents code, but not the results from appearing in the report. Use this when writing reports aimed at people who don’t want to see the underlying R code.
Use tab-completion to see available options!
Read about knitr specific options in the developer’s documentation or the Quarto Code Cells: Knitr reference.
Content adapted from the Quarto chapter of R for Data Science.
💪🏼 Exercise 1-6
Add appropriate labels to the first and last code chunks.
Try navigating to the chunks using the drop down navigator at the bottom left of the source pane.
Add
#| echo: falseto all chunks and re-render. How did this change the report?Try two more chunk options and re-render.
To see all options, type#|thenTab, orCmd/Ctrl+Space.💬 Discuss: Share which options you tried and what they did.
Use the Code Cells: Knitr reference as needed.
06:00
Global options
Use for chunk options you want to apply to all chunks by default.
Code execution options:
Some global options relevant only to R must be set under the knitr key, under opts_chunk:
Content adapted from the Quarto chapter of R for Data Science.
💪🏼 Exercise 1-7
Delete the
echo: falseoption from all code chunks.Add
code-fold: true(under thehtml:key in the YAML header) to collapse all code chunks by default.💬 Discuss: What do you think will happen if you add
echo: false(under theexecute:key in the YAML header)? Try it out.Try adding a
knitr: opts_chunk:global option. Careful with indentation!Tab-completion doesn’t seem to work for the
opts_chunkYAML key.
Use the Code Cells: Knitr reference as needed.
⏱10-min break after this exercise
06:00
🚶🏻♀ Break 🧘🏻♀️
10:00
Parameterized reports
Many use cases
Different audiences, different reports
Show code for technical staff and hide code for everyone else (StackOverflow example).
Like a custom function
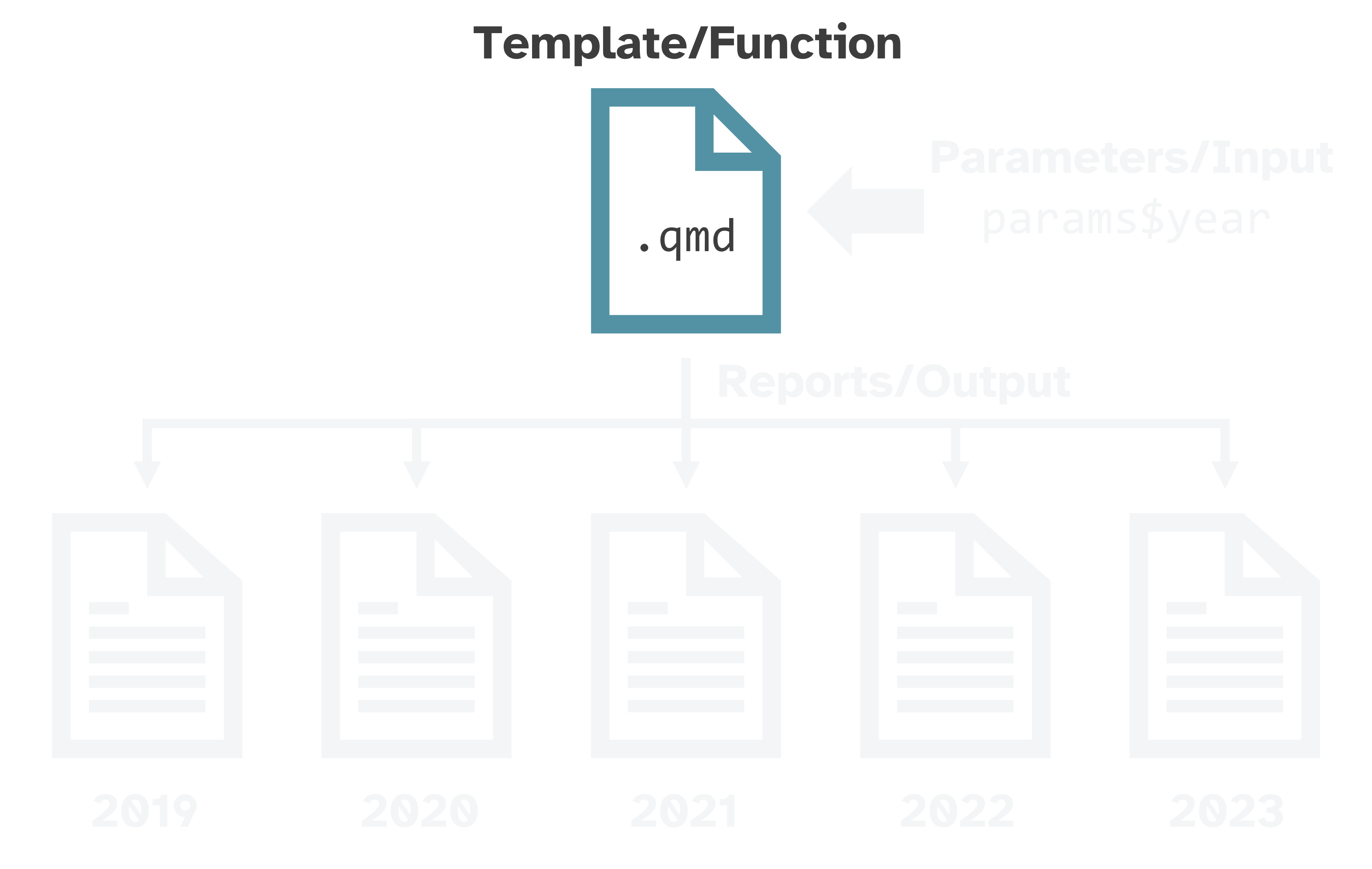
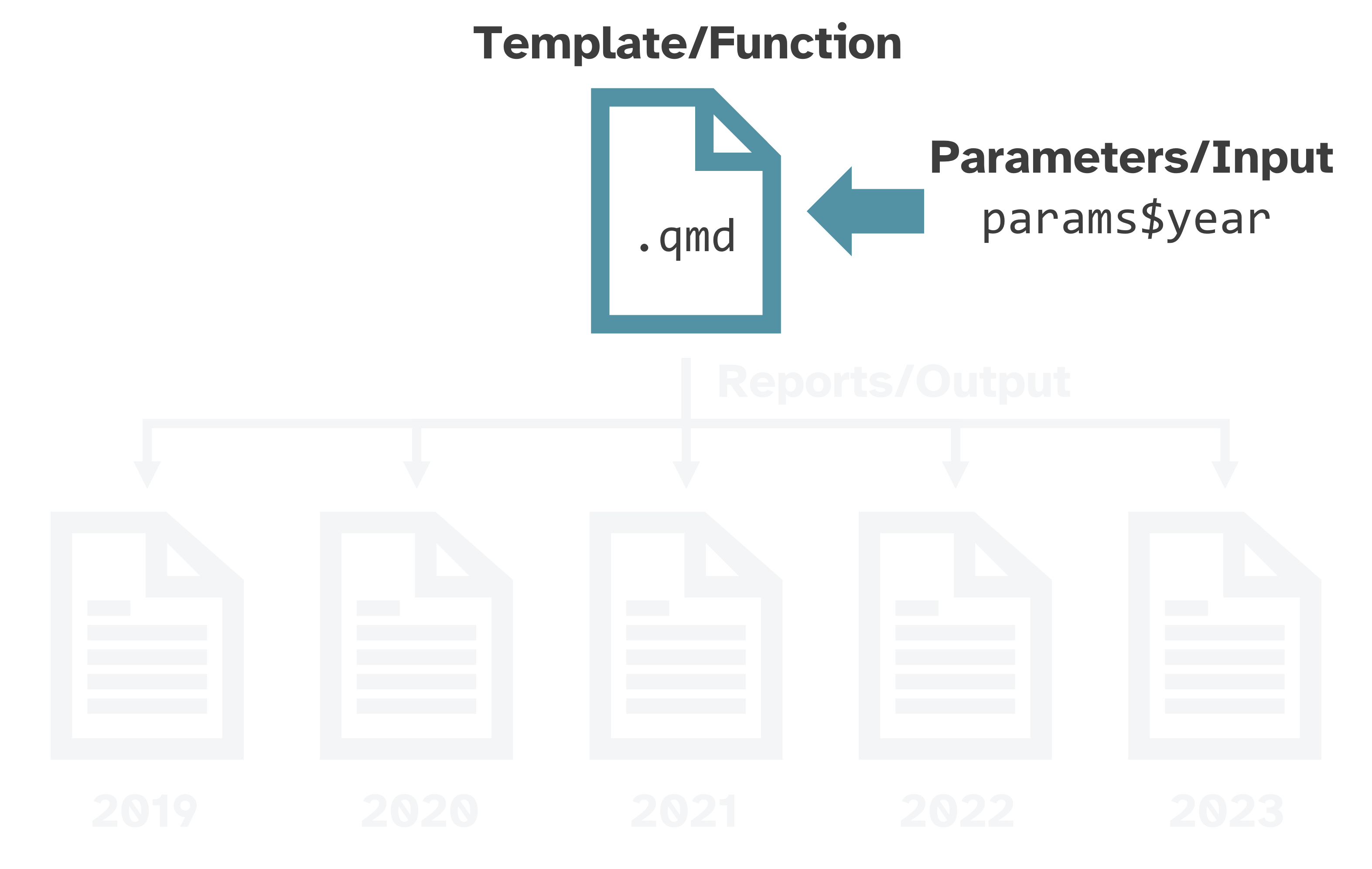
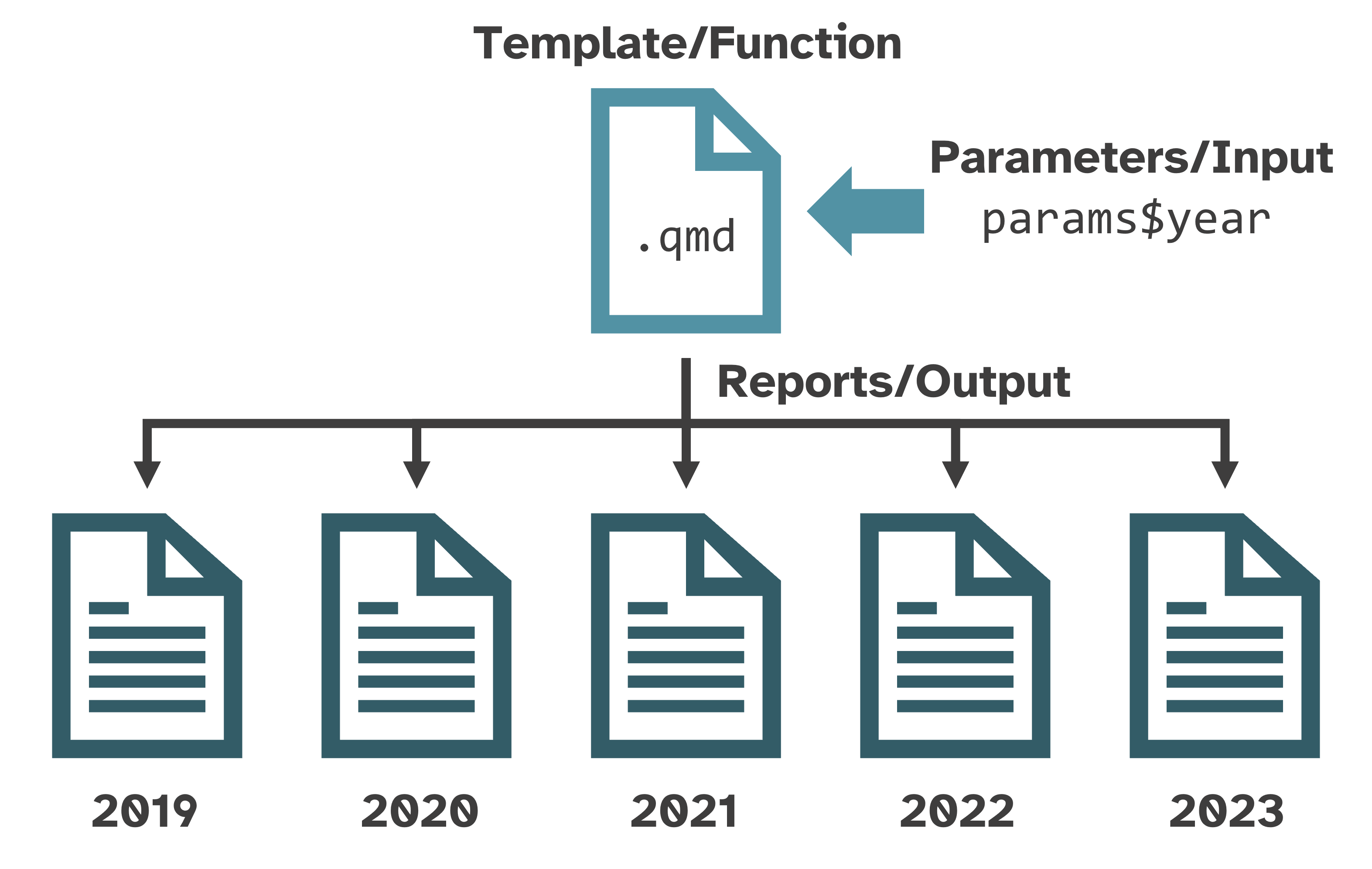
What makes a report “parameterized”?
YAML header with
paramskey-value pairsUse these
paramsto create different variations of a report from a single.qmddocument.
Important
Valid parameter values are strings, numbers, or Boolean.
Must serialize a dataframe to pass it as a parameter, then un-serialize it back to a dataframe within the
.qmdcontent.See Christophe Dervieux’s answer in Posit Community to understand why.
See John Paul Helveston’s blog post to learn how to use {jsonlite} as a workaround.
Workflow
Write report template with default values hard-coded, and then render & review.
Set default
paramskey-value pairs in YAML.Replace hard-coded values with the
paramsvariables.Render the single report and review.
Render extreme cases and review.
- Parameter values with barely any data and with tons of data.
Render all variations of the report at once.
💪🏼 Exercise 2
Explore a report without parameters and see where we could add them.
Open
2-swiss-cats.qmd.Click the
![Quarto render button in RStudio]() Render button.
Render button.Look at the source markdown & code and the rendered report.
💬 Discuss: What variables could we set as parameters?
💡 Hint: run the
setupchunk and look at thepetsdataframe to see what variables it has.
05:00
Set params in YAML header
---
title: "Swiss Cats" # Metadata
format: # Set format types
html:
toc: true # Set additional options
docx: default
params: # Set default parameter key-value pairs
fave_breed: "Snowshoe"
---
Report content goes here. # Write narrative and codeImportant
Your default params key-value pairs must be found in your dataset. Otherwise, code will error or output will be blank.
The variable name for params can be anything you choose. Often, it’s a column name in your dataset.
Access params
Run any line or chunk to add params to your environment.
Replace hard-coded values with params
Cmd/Ctrl+Fto find where to replace hard-coded values withparams.
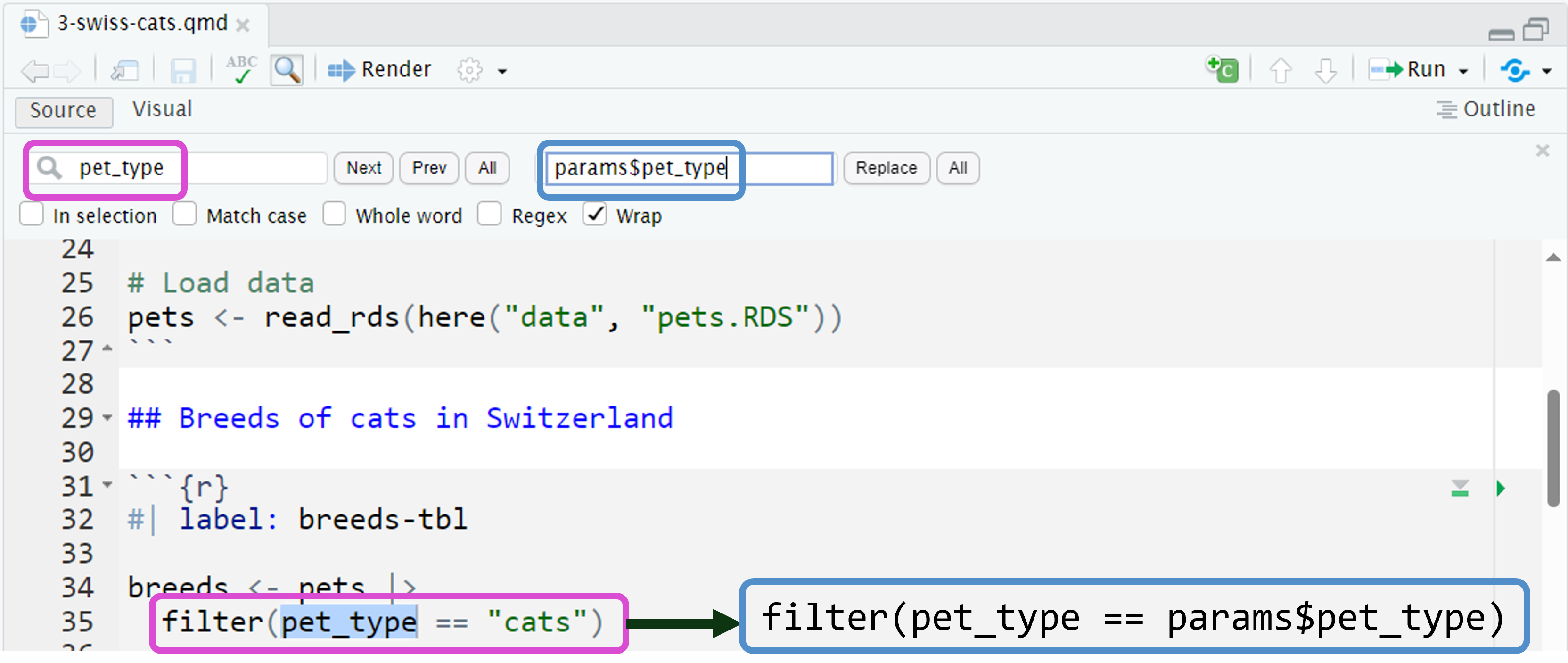
Replace hard-coded values with params
Use inline R code for markdown.
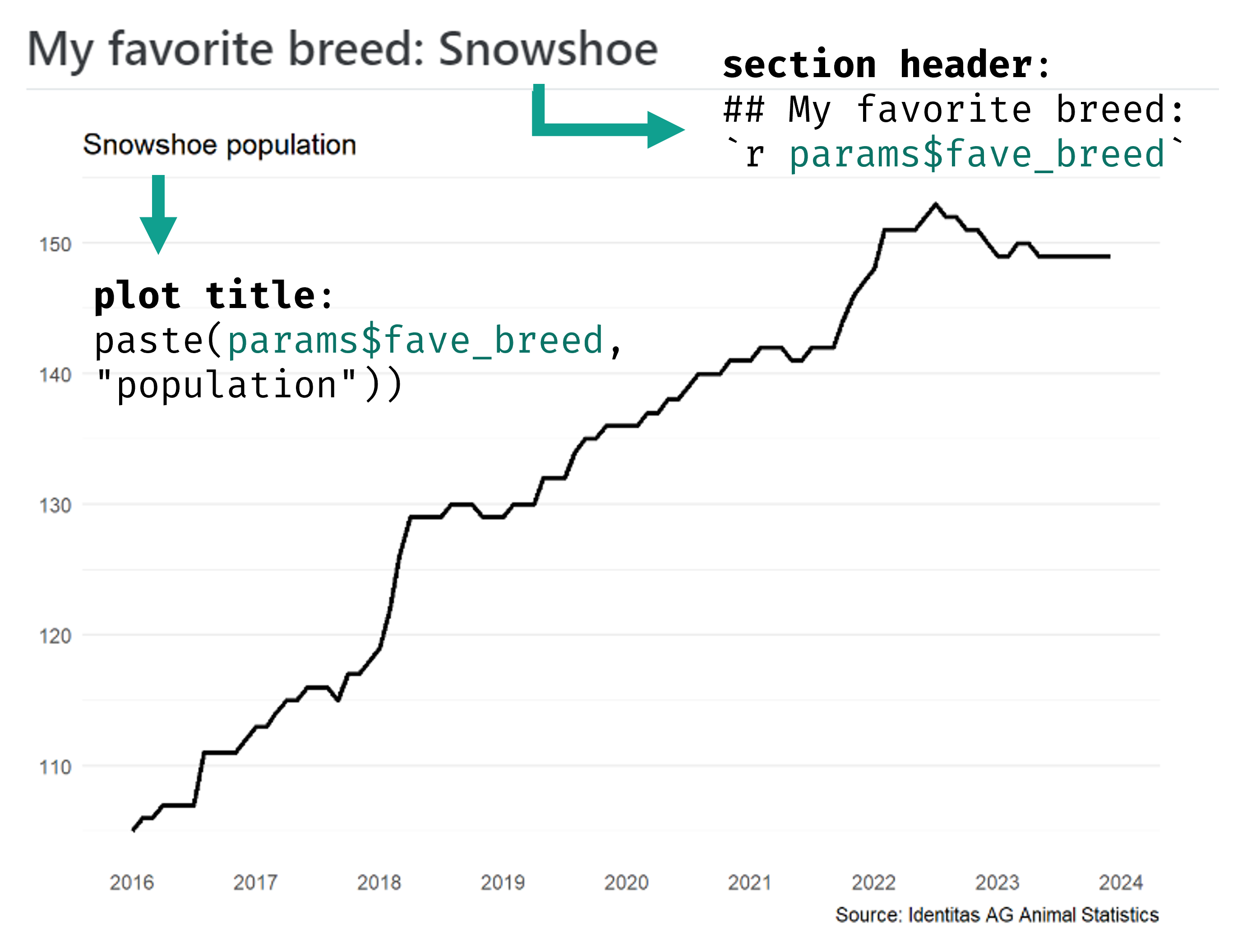
💃🏻 Demo
Modify 2-swiss-cats.qmd to add pet_type and fave_breed parameters.
This parameterized version of 2-swiss-cats.qmd is the starting point for the next section’s exercises (3-quarto-render.qmd).
Rendering many reports
Review three ways to render
RStudio/Quarto integration:
![Quarto render button in RStudio]() Render button in RStudio or
Render button in RStudio or Cmd/Ctrl+Shift+Kkeyboard shortcut✨ Quarto R package ✨
Quarto CLI
💪🏼 Exercise 3-1
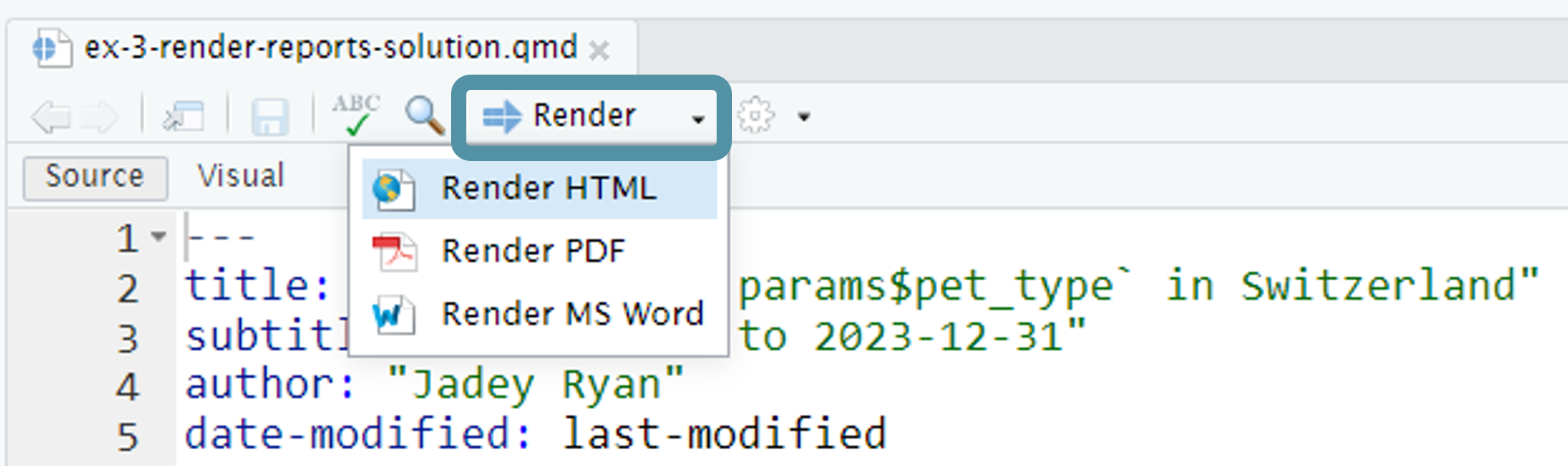
Change parameters in the YAML and render using ![]() Render button.
Render button.
Look at the unique pet breeds and pick your favorite.
In
3-quarto-render.qmdChange the default parameters in the YAML to your favorite pet type and breed. Render using the![Quarto render button in RStudio]() Render button.
Render button.💬 Discuss: Which breed did you pick and why? What do you think will happen if you set the
pet_typedefault parameter to “cats” and thefave_breedparameter to “American Bulldog”?
Try it out!
07:00
💪🏼 Exercise 3-2
Change parameters and render using quarto_render().
Render with
quarto::quarto_render().💬 Discuss: What kinds of variables will you use as parameters for your reports?
⏱10-min break after this exercise
07:00
🚶🏻♀ Break 🧘🏻♀️
10:00
Render all 538 reports
One HTML report for each cat breed and each dog breed.
Change the default
paramsin the YAML.Render button or
Cmd/Ctrl+Shift+Kkeyboard shortcut.Rename the rendered report to include the parameter & prevent overwriting.
Repeat 537 times.
😭
quarto::quarto_render(
input = here::here("3-quarto-render.qmd"),
output_file = "dogs-affenpinscher-report.html",
execute_params = list(
pet_type = "dogs",
fave_breed = "Affenpinscher"))
quarto::quarto_render(
input = here::here("3-quarto-render.qmd"),
output_file = "dogs-afghan-hound-report.html",
execute_params = list(
pet_type = "dogs",
fave_breed = "Afghan Hound"))
quarto::quarto_render(
input = here::here("3-quarto-render.qmd"),
output_file = "dogs-aidi-chien-de-montagne-de-l-atlas-report.html",
execute_params = list(
pet_type = "dogs",
fave_breed = "Aidi Chien De Montagne De L Atlas"))
quarto::quarto_render(
input = here::here("3-quarto-render.qmd"),
output_file = "dogs-akita-report.html",
execute_params = list(
pet_type = "dogs",
fave_breed = "Akita"))
# + 534 more times...
# 😭Create a dataframe with three columns that match quarto_render() args:
output_format: file type (html, revealjs, pdf, docx, etc.)output_file: file name with extensionexecute_params: named list of parameters
Map over each row:
purrr::pwalk(dataframe, quarto_render, <arguments for quarto_render>)😎
purrr map functions for iteration
Map functions apply the same action/function to each element of an object.
Base R
apply()functions are map functions.purrrmap functions have consistent syntax and the output data type is predictable.
for loops → lapply() → purrr::map()

Learn more:
R-Ladies Baltimore presentation Make your R Code purr with
purrr
Create dataframe to iterate over
pet_reports <- pets |>
dplyr::distinct(pet_type, breed) |> # Get distinct pet/breed combos
dplyr::mutate(
output_format = "html", # Make output_format column
output_file = paste( # Make output_file column:
tolower(pet_type), # cats-abyssiniane-report.html
tolower(gsub(" ", "-", breed)),
"report.html",
sep = "-"
),
execute_params = purrr::map2( # Make execute_params column
pet_type,
breed,
\(pet_type, breed) list(pet_type = pet_type, breed = breed)))Subset to first 2 cat/dog breeds
pet_reports_subset <- pet_reports |>
dplyr::slice_head(n = 2, by = pet_type) |>
dplyr::select(output_file, execute_params)
pet_reports_subset| output_file | execute_params |
|---|---|
| cats-abyssiniane-report.html | cats , Abyssiniane |
| cats-aegean-cat-report.html | cats , Aegean Cat |
| dogs-affenpinscher-report.html | dogs , Affenpinscher |
| dogs-afghan-hound-report.html | dogs , Afghan Hound |
Map over each row
purrr::pwalk()iterates over multiple arguments simultaneously.First
.largument is a list of vectors.- Dataframe is a special case of
.lthat iterates over rows.
- Dataframe is a special case of
Note
index is the only named argument of quarto_render() included in pwalk().
output_format, output_file, and execute_params are already passed in through the dataframe.
Multiple formats
Render all reports to all formats
Add to the format: YAML option to render additional output formats from the same .qmd file.
Format links for HTML output
Links to download the other formats will automatically appear in HTML documents.
Format link options
Choose which format links to include:
💃🏻 Demo
Demo programmatically rendering all reports in all formats in 4-demo-quarto-render-purrr.qmd and 4-demo-quarto-render-purrr.R.
Watchouts
Can’t render reports to another directory.
output-dirYAML option only works for Quarto projects that contain a_quarto.ymlconfig file.Workaround: use
{fs}to move files after rendering.See
demo-quarto-render-purrr.Rfor example.More info: GitHub discussion and GitHub issue.
If using
embed-resources: trueYAML option,.qmdcan’t be in subfolder, otherwise:[WARNING] Could not fetch resource …
More info: GitHub discussion and GitHub issue.
Conditional code execution
Conditionally execute a code chunk
More efficient to not execute code that generates interactive outputs for static reports.
Useful for executing interactive plot code (e.g.,
plotlyorggiraph) for HTML reports and staticggplot2code for all other formats.Useful for executing different code based on a parameter value.
Not currently a feature of Quarto v1.4. Follow along with this GitHub discussion.
Chunk options can use R code for option values with
!expr. Learn about the limitations to this YAML “tag” literal syntax in the Quarto Chunk Options reference.
Conditional code based on output
Include in the setup chunk of your .qmd file.
Get the format of the Pandoc output:
Use eval: !expr chunk option
💪🏼 Exercise 5
Conditionally execute ggplot2 code for static reports & plotly code for interactive reports.
Open
ex-5-conditional-code.qmd.Fill in the blanks for the
eval:option forggplotcode chunks andplotlycode chunks.💬 Discuss: How would you change the
eval:option to execute a chunk based on a parameter value rather than the output format?
05:00
Conditional code based on parameter
Use params in !expr:
#| eval: !expr !params$fave_breed == "Snowshoe"
# Code for a different plot for all other breeds.
# Note the ! in front of params.Community project: home energy monitoring
Consider using a different conditional code chunk to create different visualizations depending on heating source.
Child documents
Generate dynamic contents with a “child” template
knitr::knit_child()andknitr::knit_expand()are functions from R Markdown that also work with Quarto“child” document is a template, which is run with different parameters
“main” document includes the output from all the different iterations of the child document
Community project: water quality monitoring monitoring
Consider creating a main document using the river as the parameter and a child document as a template for the heading, plots, tables, and text for each sampling site.
Do not run interactively!
Code chunks including knitr::knit_child() are not supposed to be used interactively (see this Stack Overflow response from the knitr developer Yihui Xie). If run interactively, you will get an error similar to the one shown below.
Error in `purrr::map_chr()`:
ℹ In index: 1.
Caused by error in `setwd()`:
! character argument expectedRead a description of this issue in WSDA’s {soils} R package vignette.
Learn more about child documents
Qiushi Yan’s blog post Generating dynamic contents in R Markdown and Quarto
Yihui Xie’s R Markdown Cookbook sections on
knitr::knit_child()andknitr::knit_expand()Isabella Velásquez’s Getting started with report writing using Quarto at 26:00.
💃🏻 Demo
Generate a report that dynamically creates sections and plots for all breeds of the pet_type.
Main document: 6-demo-knit-child.qmd
Child document: _child-template.qmd
🏁 Recap
Learning objective 1
Learn what Quarto is and what you can use it for.
Scientific and technical publishing system for:
- interactive HTML reports
- static MS Word or PDF fact sheets or reports
- manuscripts
- presentations
- dashboards
- websites
- books
- blogs
Check out the Quarto Gallery!

Learning objective 2
Learn how to weave code and text together to create a fully reproducible report.
.qmd documents contain:
- document level metadata and options in the YAML
- a narrative using markdown syntax, and
- code chunks to import, wrangle, visualize data.
Learning objective 3
Learn how to use parameters to create variations of a report.
Parameterized reports → very fancy custom functions:
Function →
.qmdtemplateInput → parameters
Output → rendered reports
Useful for creating variations of the same report:
spatial: country, state, county, or city
temporal: year or other time period
water bodies, sampling sites, energy sources, breeds, species, diseases, trials, etc.
Note
We only covered reports, but you can also parameterize revealjs presentations! See this Jumping Rivers blog post about it.
Thank you!
🏡 Home for all workshop materials: jadeyryan.quarto.pub/ceds-quarto-workshop/
🎥 Recordings from previous workshops & talks:
links in GitHub repo or my YouTube playlist


Reproducible Reporting with Quarto // jadeyryan.quarto.pub/ceds-quarto-workshop/